Carleton University Site
ROOT
The framework of QSigEx is based on the CERN Object-Oriented data analysis
tool ROOT.
The 1-D PDF's are loaded in ROOT (THF1) and all the correlations can be calculated while the template
PDF's are prepared (TTree). Event pre-selection can be apply while producing the PDF's.
All the PDFs, cuts, and fitted results can be viewed in ROOT.
The ROOT interface provides all the info to be stored in TObject and TDirectory. The user communicate
with QSigEx with a CARDFILE such that the QSigEx class instances can be created:
QSigExDirHandler: Abstract base class for TDirectory handler classes
QSigExCuts: Loads equivalences/cuts in the TDirectory structure; treats regular 1D selections
and/or expressions
QSigExCleanData: Applies cuts on raw data events and fills a TTree with clean data
QSigExProbs: Computes the marginal probability densities using marginal PDFs and data TTtree
QSigExPDFs: Abstract base class for QSigExDirHandler derived classes that load marginal PDFs in
the TDirectory structure
QSigExJointProbs: Computes the joint probability densities of variables with/without
correlation (full or multi-dim)
QSigExFit: Evaluates the population parameters and their errors using Minuit minimization
package (binned/unbinned/analytic)
Fit the pure D2O Data Sample
Here is a full example on how to compute the published fluxes for the pure D2O data sample.
Results were reported in PRL.
The following instructions assume QSIGEX is installed
on your system.
1) Go into the directory QSIGEXDIR/runme/d2o
> cd $QSIGEXDIR/runme/d2o
2) Compile the executable (rund2o)
> make
3) Run It!
> rund2o d2o_pdfs.dat d2o_results.root
Signal PDF's (signal_pdf.root)
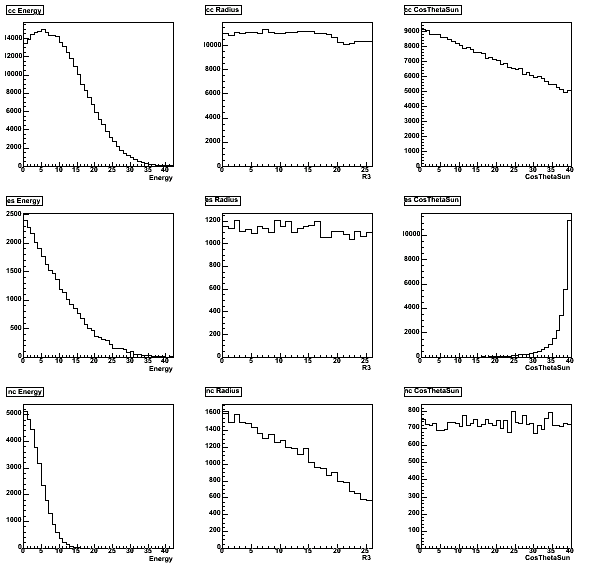
The marginal 1D PDF's for the three fluxes: energy E [42 bins] - radius R3 [26 bins] - cosThetaSun [40 bins].
The mapping corresponds to energy [5.511,16.011 MeV] - radius [0,166.375e6 cm3] - cosThetaSun [-1,+1].
This root file can be found in $QSIGEXDIR/runme/d2o.
Background PDF's (bck_pdf.root)
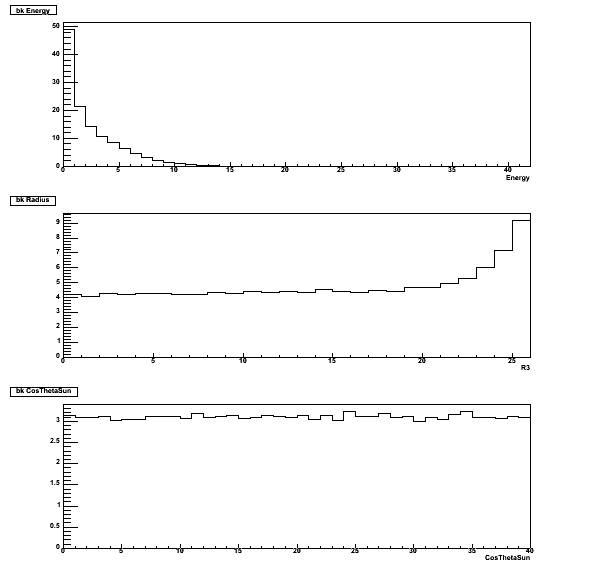
The marginal 1D PDF's for the three fluxes: energy E [42 bins] - radius R3 [26 bins] - cosThetaSun [40 bins].
The mapping corresponds to energy [5.511,16.011 MeV] - radius [0,166.375e6 cm3] - cosThetaSun [-1,+1].
The background PDF is composed of 51.7 internal p-d neutrons, 27 external neutrons, 20.4 D2O beta-gammas,
16 PMT beta-gammas, and 9.1 AV beta-gammas.
This root file can be found in $QSIGEXDIR/runme/d2o.
The Data (d2o_data.root)
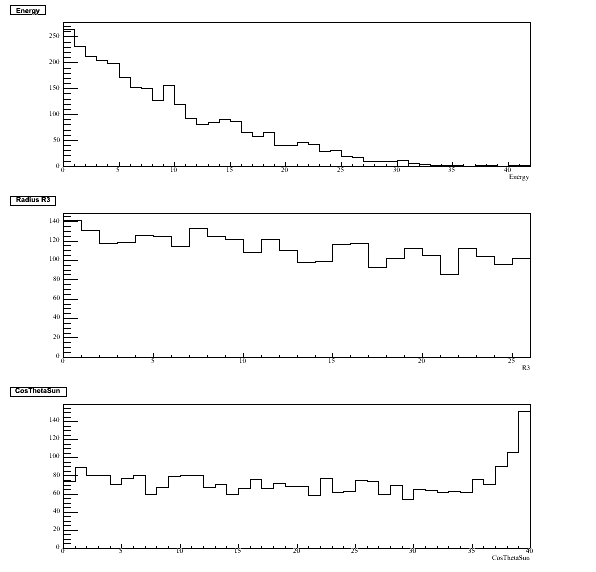
The marginal 1D PDF's for the three fluxes: energy E [42 bins] - radius R3 [26 bins] - cosThetaSun [40 bins].
The mapping corresponds to energy [5.511,16.011 MeV] - radius [0,166.375e6 cm3] - cosThetaSun [-1,+1].
This root file can be found in $QSIGEXDIR/runme/d2o.
Understand the CARDFILE (d2o_pdfs.dat)
DATA_FILE d2o_data.root Tree
Name of the ROOT file for the data and of the TTree identifier (i.e. Tree)
# name coordinate condition value
Pre-selection cuts
cut emin energy>=0
cut emax energy<=42
cut rmin radius>=0
cut rmax radius<=26
cut cmin cthsun>=0
cut cmax cthsun<=40
# flag class flux_group dimension name PDF filename data field NBins
pdf 1 TH1F cc syste h1001 signal_pdf.root energy
TH1F 1D histogram - cc group - systematic class - ID h1001 - ROOT file - variable (energy)
pdf 1 TH1F cc systr h1002 signal_pdf.root radius
TH1F 1D histogram - cc group - systematic class - ID h1002 - ROOT file - variable (radius)
pdf 1 TH1F cc systc h1003 signal_pdf.root cthsun
TH1F 1D histogram - cc group - systematic class - ID h1003 - ROOT file - variable (cthsun)
pdf 1 TH1F es syste h2001 signal_pdf.root energy
TH1F 1D histogram - es group - systematic class - ID h2001 - ROOT file - variable (energy)
pdf 1 TH1F es systr h2002 signal_pdf.root radius
TH1F 1D histogram - es group - systematic class - ID h2002 - ROOT file - variable (radius)
pdf 1 TH1F es systc h2003 signal_pdf.root cthsun
TH1F 1D histogram - es group - systematic class - ID h2003 - ROOT file - variable (cthsun)
pdf 1 TH1F nc syste h3001 signal_pdf.root energy
TH1F 1D histogram - nc group - systematic class - ID h3001 - ROOT file - variable (energy)
pdf 1 TH1F nc systr h3002 signal_pdf.root radius
TH1F 1D histogram - nc group - systematic class - ID h3002 - ROOT file - variable (radius)
pdf 1 TH1F nc systc h3003 signal_pdf.root cthsun
TH1F 1D histogram - nc group - systematic class - ID h3003 - ROOT file - variable (cthsun)
pdf 1 TH1F bk syste h4001 bck_pdf.root energy
TH1F 1D histogram - bk group - systematic class - ID h4001 - ROOT file - variable (energy)
pdf 1 TH1F bk systr h4002 bck_pdf.root radius
TH1F 1D histogram - bk group - systematic class - ID h4002 - ROOT file - variable (radius)
pdf 1 TH1F bk systc h4003 bck_pdf.root cthsun
TH1F 1D histogram - bk group - systematic class - ID h4003 - ROOT file - variable (cthsun)
# parameter_name active(1) startval minval maxval stepval
flux cc 1 2000 0 10000 0.01
The starting/minimum/maximum values for the cc flux and the precision
flux es 1 200 0 1000 0.01
The starting/minimum/maximum values for the es flux and the precision
flux nc 1 500 0 1000 0.01
The starting/minimum/maximum values for the nc flux and the precision
flux bk 1 124.2 124.2 124.2 0.00
The starting/minimum/maximum values for the bk flux and the precision
#Minuit control
minimizer MIGrad 1
Invoke Minuit MIGrad interactive command (1 sigma)
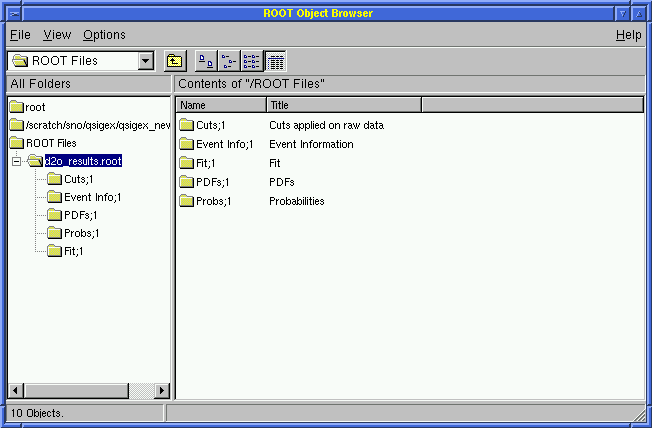
Open a TBrowser in ROOT!
Use the ROOT executable in $QSNO_ROOT/bin/root
> root d2o_results.root
root [1] TBrowser a;
Set the View at Details (the default is Small Icons)
Click on ROOT File
Click on d2o_results.root
The user will see:
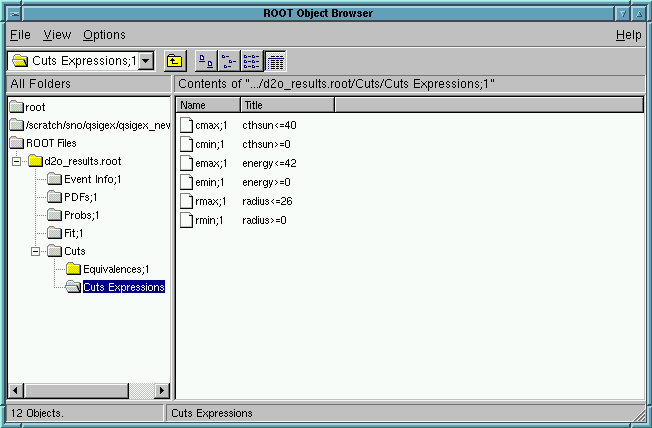
View the Cuts in ROOT!
Click on Cuts and then on Cuts Expressions
The user will see:
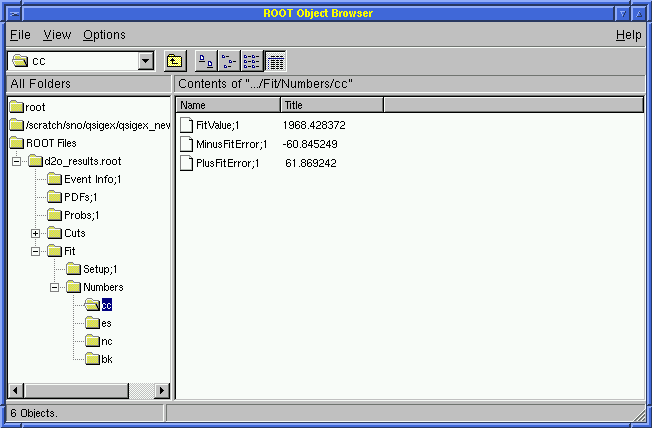
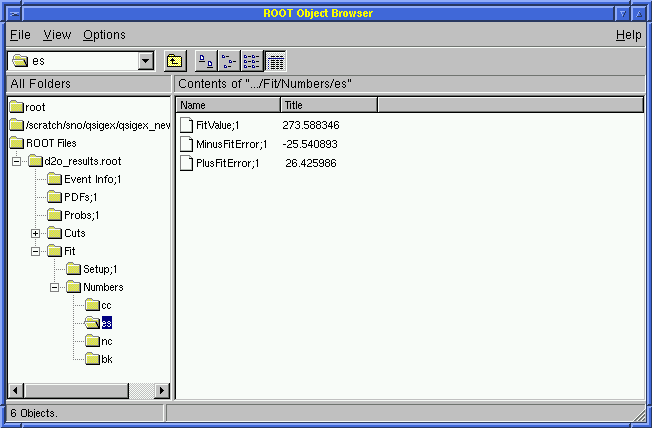
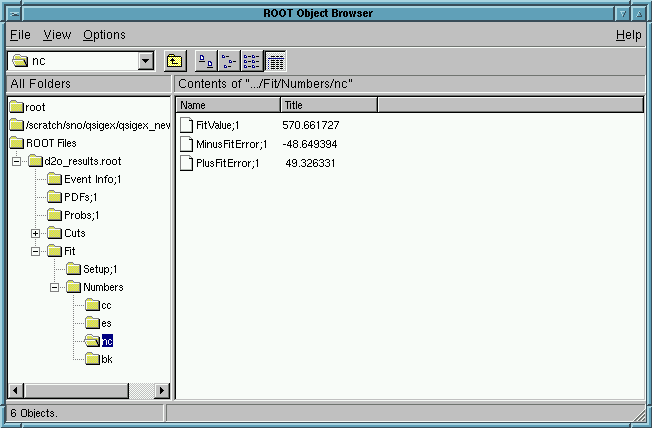
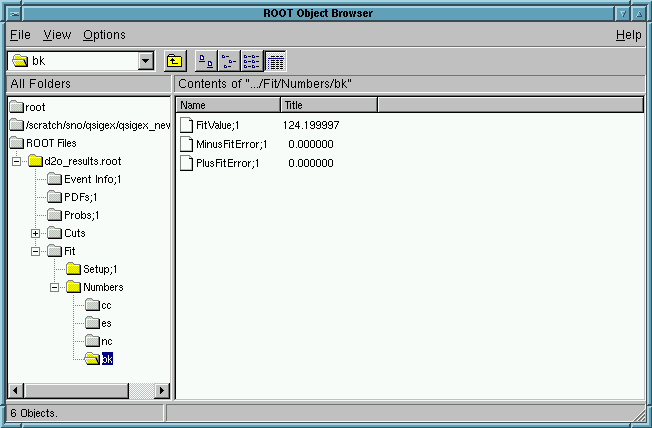
View the Results for the fitted fluxes in ROOT!
Click on Fits and then on Numbers
Click on cc or es or nc or bk
The user will see the CC, ES, and NC fluxes as well as the background (fixed):
Compare QSigEx and the UNIDOC
The QSigEx results presented above compared to the standard FORTAN
code (sigex.f) from the UNIDOC. Both code uses the same
data and the same PDF's. Hence they give consistent results. QSIGEX uses a PDF
for the background; while the FORTRAN code
subtract the background for the data. This explains the differences for the last
digits (especially for the NC flux).
|
QSIGEX | UNIDOC |
|
2936 [Data] | 2936 [Data] |
|
1968.4 +61.9 -60.8 [CC] | 1968.5 +61.8 -60.8 [CC] |
|
273.6 +26.4 -25.5 [ES] | 273.2 +26.4 -25.6 [ES] |
|
570.7 +49.3 -48.7 [NC] | 572.7 +49.3 -48.6 [NC] |
|
124.2 (fixed) [BK] | 124.2 (subtracted) [BK] |
|
2936.9 [Total Fitted] | 2938.6 [Total Fitted] |
The correlations between the fluxes are the same.
|
QSIGEX | UNIDOC |
|
-0.516 [CC-NC] | -0.516 [CC-NC] |
|
-0.157 [CC-ES] | -0.157 [CC-ES] |